
26 Sep. 2022
NeuroMiner now allows for individual prediction result interpretation through the visualisation window.
Click here to learn more
26 Sep. 2022
NeuroMiner now allows you to estimate the performance of models trained with simulated data
in order to find out the optimal sample size needed for your analysis.
Click here to learn more
NeuroMiner is for free!
We fully endorse the principles of open software and open science to accelerate urgently needed progress in the treatment of complex brain disorders.
Use our software for free if you work in academia or obtain a license if you want to use NeuroMiner for commercial purposes.
Powerful machine learning methods at your fingertips!
Do not spend your time on learning how to code or how to integrate different machine learning libraries.
Make most of your data!
Flexible data and strategy fusion methods enable you to extract predictive information across different data domains.
Thus you can build more comprehensive models for a better management of complex system-level disorders.
Speed up your ML computations!
Users with access to high-performance computing can massively accelerate NeuroMiner computations by parallelizing our software.
Be on the safe side!
Robust nested cross-validation allows you to test the
generalizability of your models in a methodologically unbiased way.
Better understand your models!
Insight into the model generation process and the predictive features it produces is
key for generating trust in your models and advance our understanding of complex system-level disorders.


NeuroMiner comes as free software to facilitate research into better tools for precision medicine.
It is constantly updated by the Section for Neurodiagnostic Applications (SNAP) in Psychiatry at the Department of Psychiatry and Psychiatry of Ludwig-Maximilian-University.
NeuroMiner requires no coding experience as it is fully menu-driven.
The main functionality of the app is easily accessible through text-based menus in MATLAB.
This allows using NeuroMiner in server/remote environments with no or limited graphical interfaces.
Furthermore, the NeuroMiner interface facilitates standardized parameter setup, storage, and dissemination across research labs,
which is an essential requirement for more robust and generalizable predictive models.
For feedback or inquiries concerning a commercial license for NeuroMiner,
Click here to get in touch with us!
NeuroMiner provides a wealth of state-of-the-art supervised Machine Learning techniques,
such as linear and non-linear support-vector machines, relevance vector machines, random forests, and gradient-boosting algorithms.
It also comes with numerous dimensionality reduction methods and feature selection strategies that allow finding
optimal combinations of predictive features for the user’s given prediction problem.
NeuroMiner can analyze all tabular data format that are stored in numeric MATLAB format,
or alternatively in CSV or Microsoft Excel spreadsheets.
It supports the in-depth analysis of 3D voxel-based neuroimaging data.
These different data sources can be analyzed separately or be combined using a variety of data fusion approaches ranging from data concatenation,
over bagging to more advanced stacking, or sequential data integration techniques.
In high-performance computing environments,
researchers can exploit the SGE-/SLURM-based parallelization functionality of NeuroMiner to tremendously speed up the model training,
cross-validation and visualization procedures.
All NeuroMiner machine learning pipelines strictly follow the principle of nested cross-validation (CV).
This is not limited to the model training phase but also includes pre-processing steps, such as scaling, standardization, or dimensionality reduction.
Thus, the inner CV cycle serves the optimization of ALL free model parameters, while the outer cycle produces methodologically unbiased estimates of generalisability.
The user can extend nested CV to repeated, nested CV to improve model robustness using ensemble learning principles.
Measures of geographic model generalisability can be produced using leave-one-site-out CV in multi-site data.
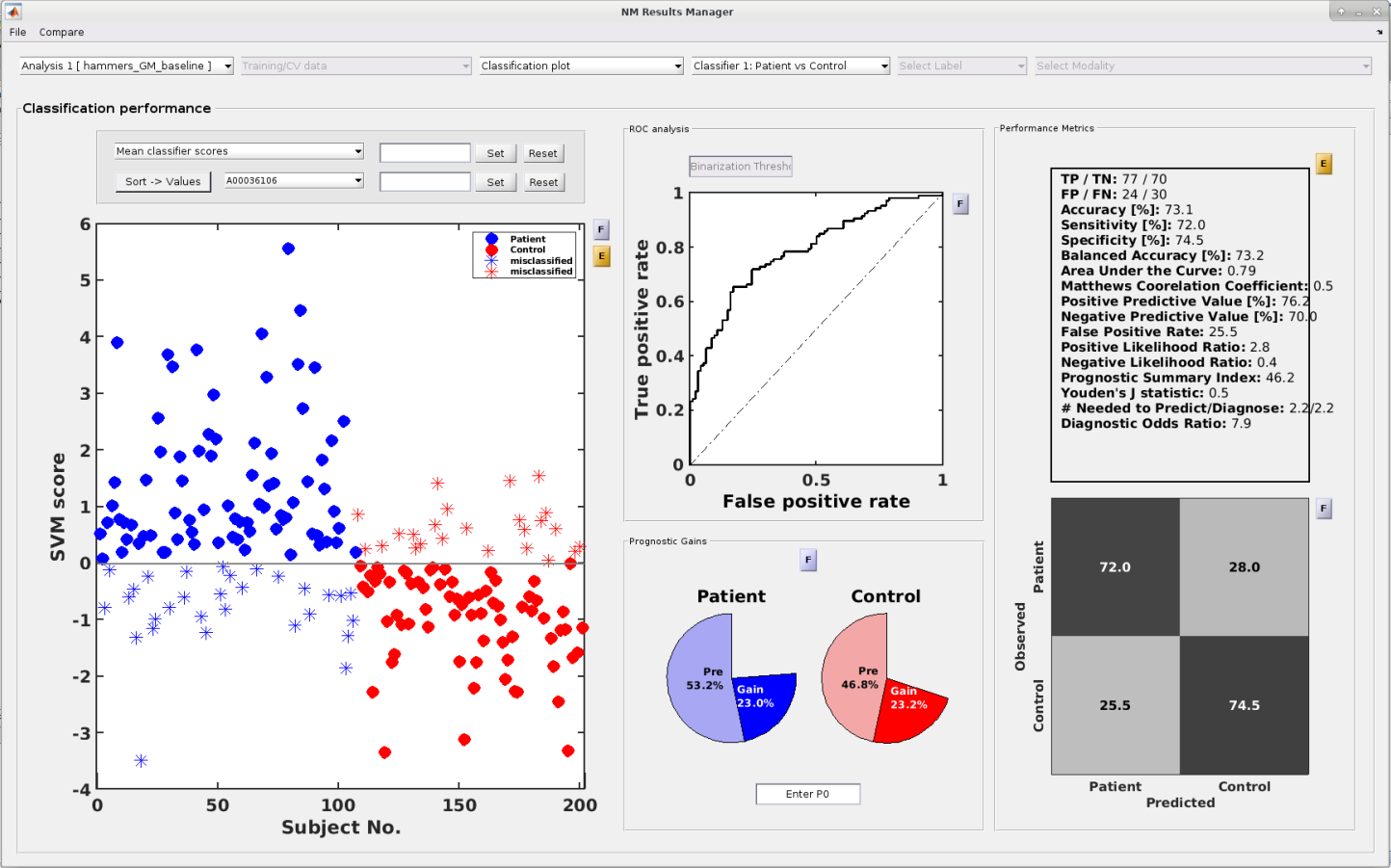
Inspection of model performance,
model optimization and predictive feature spaces are key to build clinically useful models and in turn advance both clinical neuroscience and clinical practice.
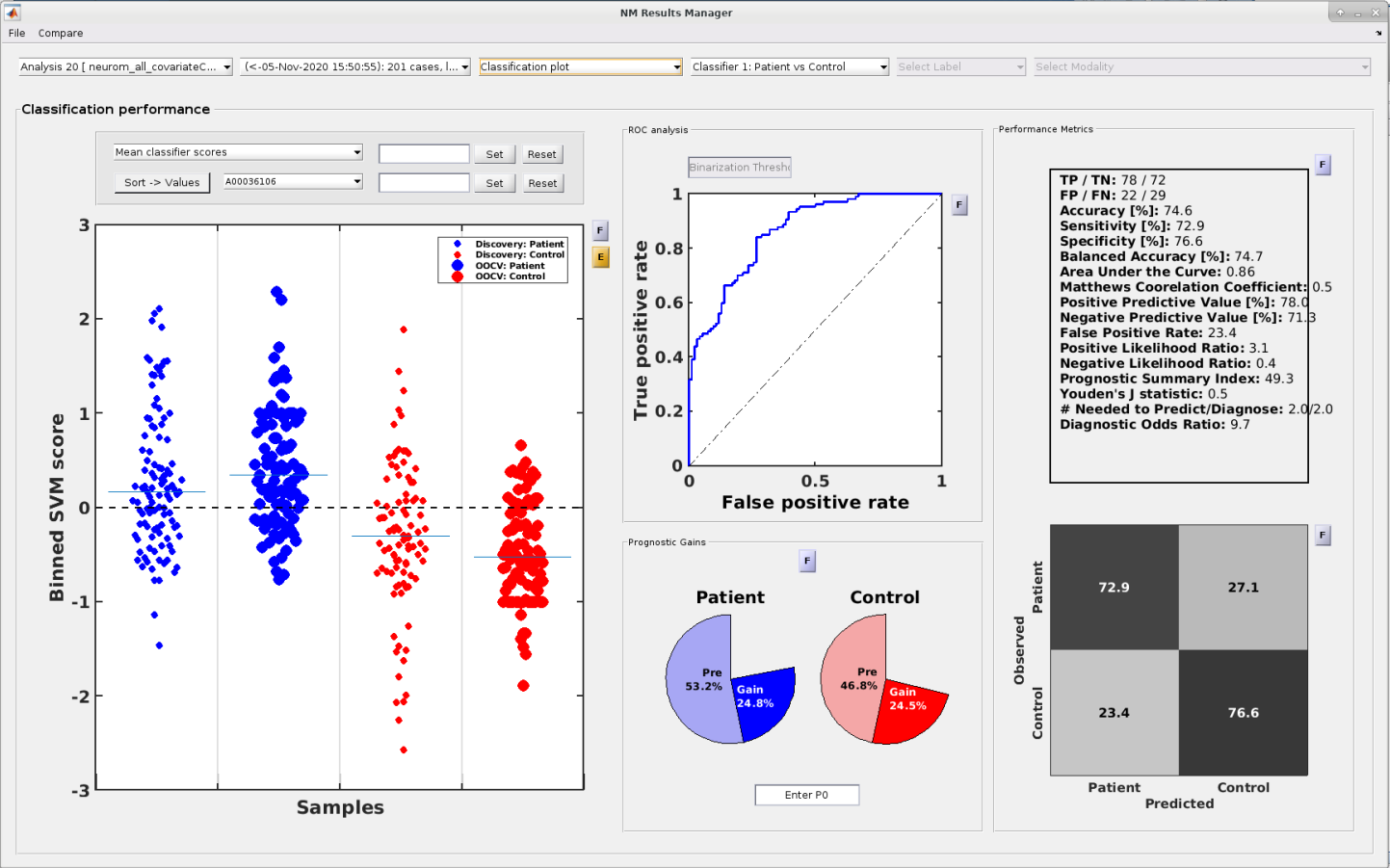
NeuroMiner supports these tasks through an intuitive graphical user interface that displays cross-validated and external validation results
as well as model performance using a bunch of performance metrics.
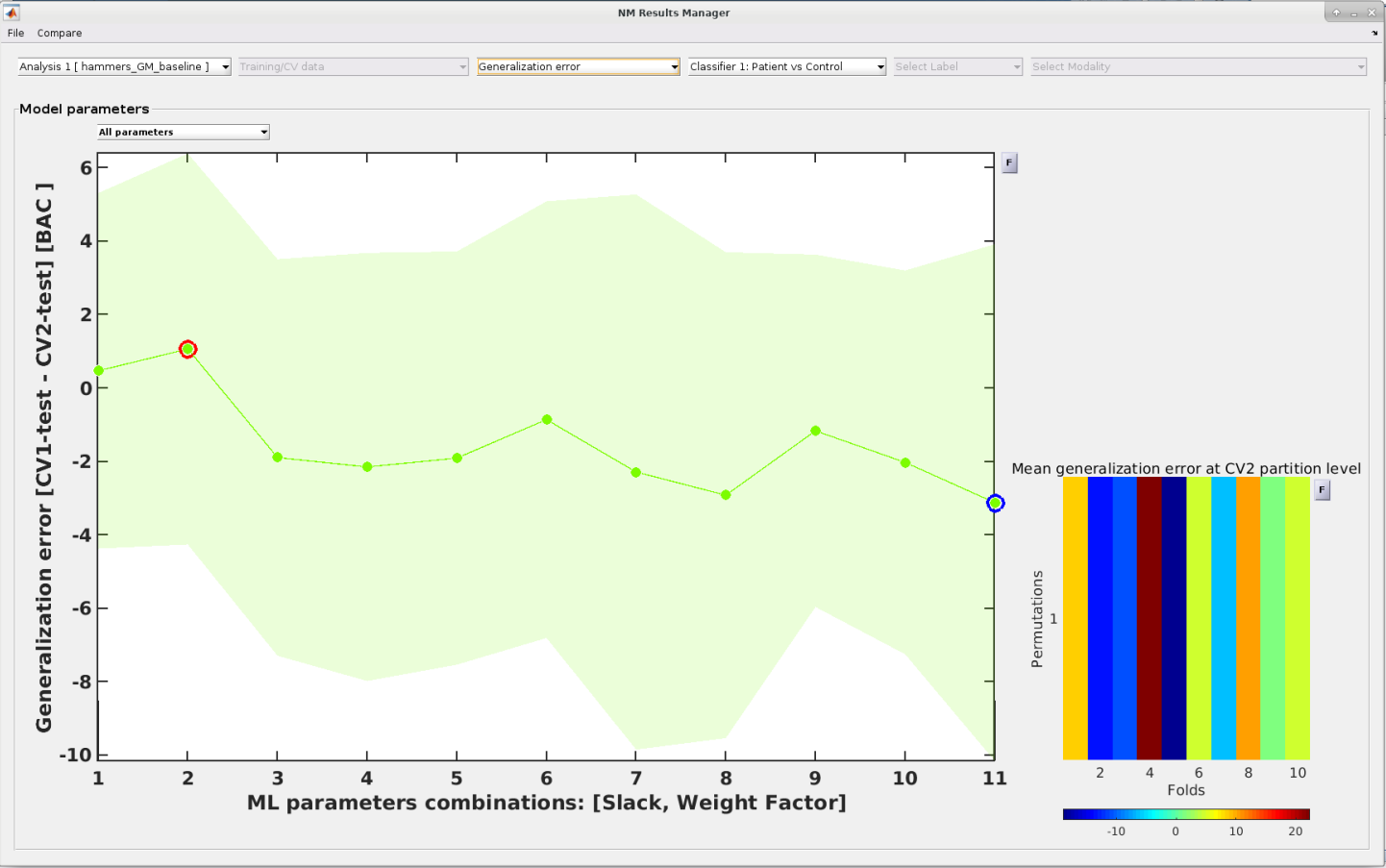
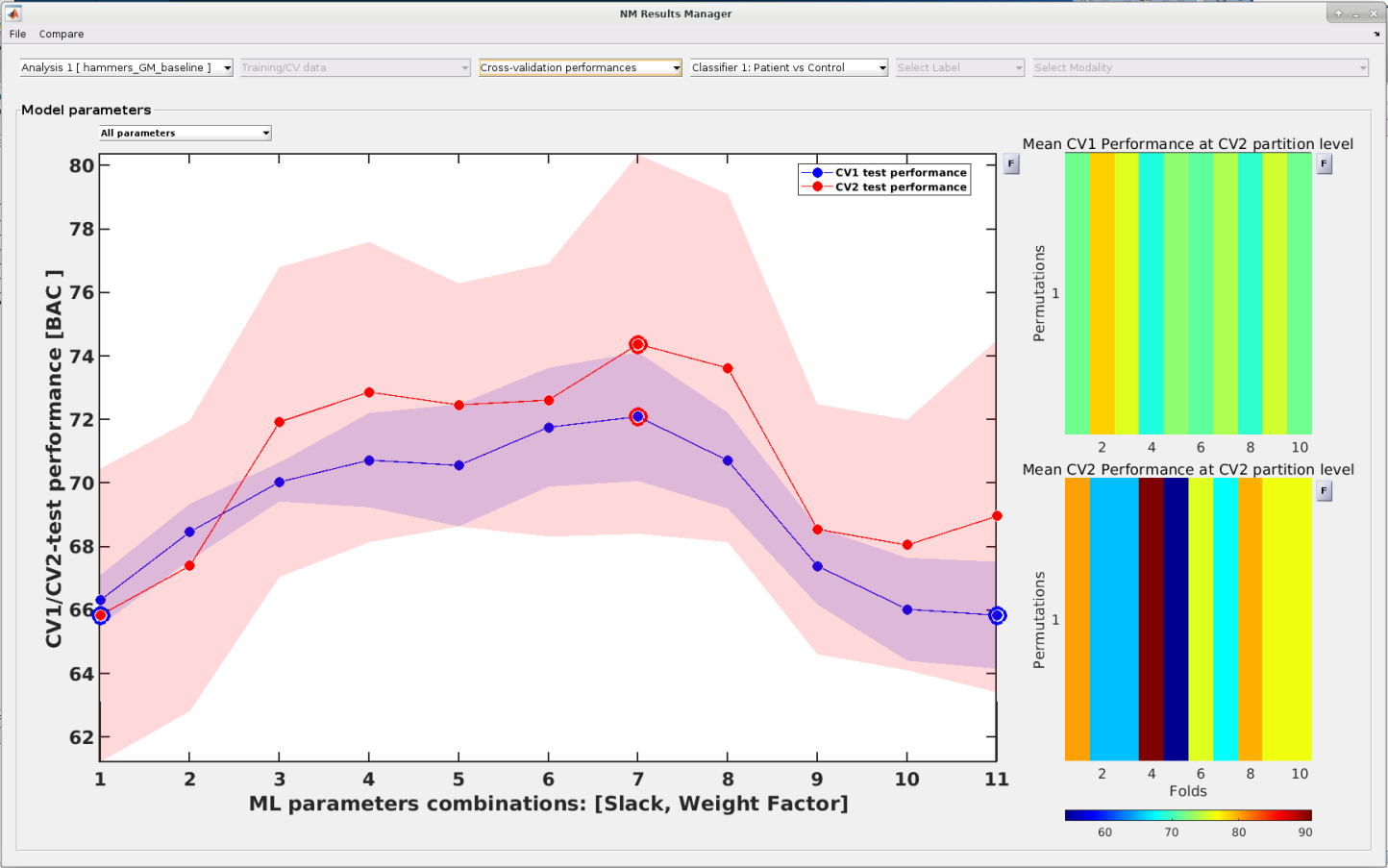
The user can also inspect the model performance metrics across the full parameter space and thus detect potential issues with their modelling strategy that
e.g. led to model overfitting.
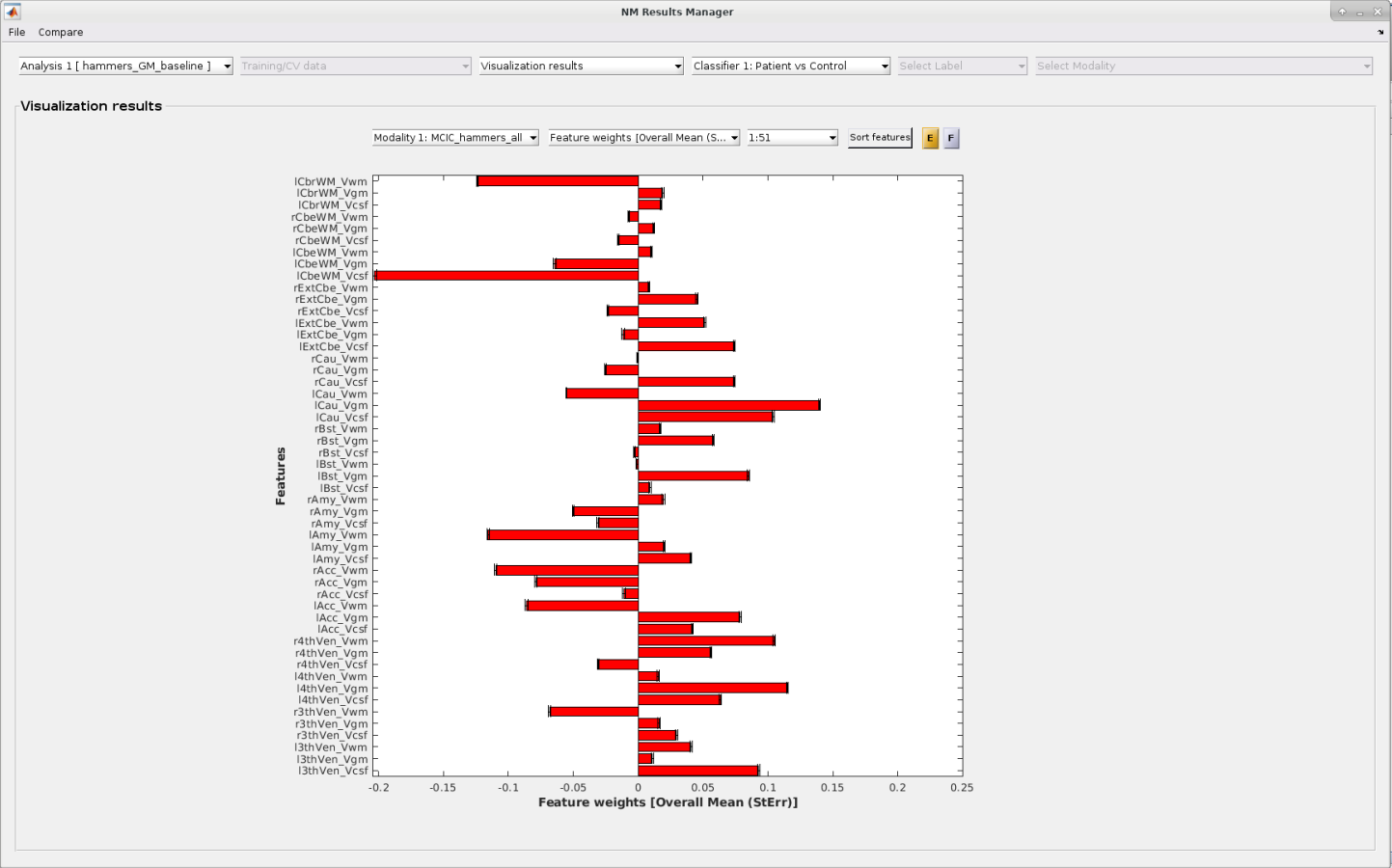
Finally, NeuroMiner can display an array of metrics of feature relevance ranging from feature selection probability,
over feature stability to measures of multivariate feature and overall model significance.
All these metrics can be easily exported into formats usable for scientific presentations and publications.
A key step in clinically relevant machine learning is the demonstration of model reproducibility/generalisability in independent patient samples.
This frequently requires the respective models to be shared across different groups of researchers.
NeuroMiner fully supports this process through dedicated external validation and anonymous data sharing modes.
Upon model completion of the model training and cross-validation phase, the researcher can enter these modes which allow them to:
1. Apply the finalized models to new datasets
2. Replace the original data with random information to safely share models without leaking sensitive information










































 L
L








 P
P L
L










 ?
?
 ?
?

















 L
L ?
?
























Legend
OSS: open source software
PEERREV: evidence of peer reviewed papers using the tool
MEDICAL: designed to be used with medical data
DEEP: deep learning implemented
S-CV: strong cross-validation design implemented as a standard (e.g., nested CV, leave-one-site-out CV)
MRI: magnetic resonance imaging
RESULTSVIS: results display showing CV results, prediction results, and display of model parameters
EXTEN.: extensibility using scripts and/or modules
DEVCOM: developer community
TEMPLATES: pipeline templates interchangeable
LIBRARY: model library built
Translational NPP: translational neurology, psychology, and psychiatry
L: limited feature realisation
P: feature achieved through external plug-in
?: unclear whether feature is present